Sorting and Layouting¶
Synopsis¶
Pangenome graphs built from raw sets of alignments may have complex structures which can introduce difficulty in
downstream analyses, visualization, mapping, and interpretation. Graph sorting aims to find the best node order for
a 1D and 2D layout to simplify these complex regions. Pangenome graphs embed linear pangenomic sequences as paths in
the graph, but to our knowledge, no algorithm takes into account this biological information in the sorting. Moreover,
existing 2D layout methods struggle to deal with large graphs. odgi implements a new layout algorithm to simplify a pangenome
graph, by using path-guided stochastic gradient descent
(PG-SGD) to move a single pair of nodes at a time.
The PG-SGD is memory polite, because it uses a path index, a strict subset of the xg index. Following a parallelized, lock-free SGD approach,
the PG-SGD can go Hogwild!
The 1D path-guided SGD implementation is a key step in general pangenome analyses such as pangenome graph linearization and simplification. It is applied in the PangenomeGraph Builder (PGGB) pipeline.
This tutorial shows how to sort and visualize a graph in 1D. It explains how to generate a 2D layout of a graph, and how to take a look at the calculated layout using static and interactive tools.
Note
Be aware that odgi sort offers much more sorting algorithms than this tutorial could cover here.
Steps¶
Build the unsorted DRB1-3123 graph¶
Assuming that your current working directory is the root of the odgi project, to construct an odgi graph from the
DRB1-3123 dataset in GFA format, execute:
odgi build -g test/DRB1-3123_unsorted.gfa -o DRB1-3123_unsorted.og
The command creates a file called DRB1-3123_unsorted.og, which contains the input graph in odgi format. This graph contains
12 ALT sequences of the HLA-DRB1 gene from the GRCh38 reference genome.
The graph was initially created with the seqwish variation graph inducer which produces unsorted, raw graphs from
all versus all alignments of the input sequences.
Visualize the unsorted DRB1-3123 graph¶
odgi viz -i DRB1-3123_unsorted.og -o DRB1-3123_unsorted.png
To obtain the following PNG image:
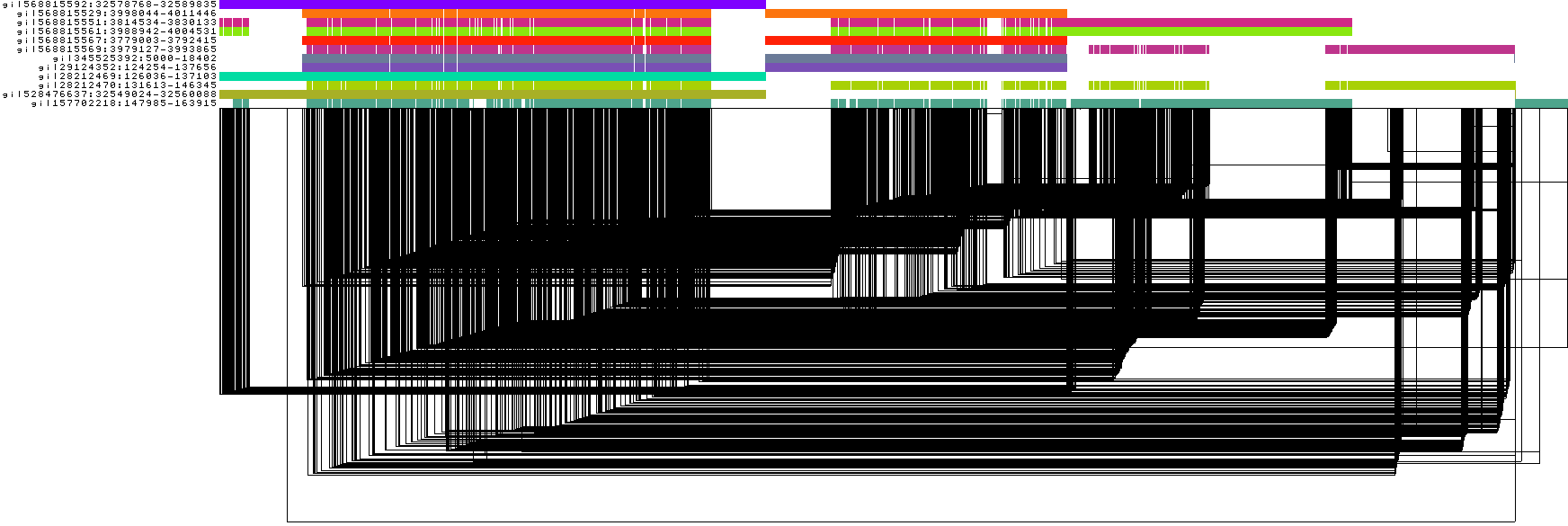
In this 1-Dimensional visualization:
The graph nodes are arranged from left to right, forming the
pangenome sequence.The colored bars represent the binned, linearized renderings of the embedded paths versus this
pangenome sequencein a binary matrix.The path names are visualized on the left.
The black lines under the paths are the links, which represent the graph topology.
The graph is very complex and, in this form, the underlying structure is unclear. It is hard to reason from it. It is a perfect candidate for the 1D PG-SGD: It will find the best 1D node order effectively linearizing it.
1D layout metrics of the unsorted DRB1-3123 graph¶
odgi stats provides metrics to evaluate the goodness of the sort of a variation graph. Let’s take a look:
odgi stats -i DRB1-3123_unsorted.og -s -d -l -g
Where:
-scalculates the sum of path node distances.-lcalculates the mean links length.-dadditionally penalizes links which connect nodes with different orientation.-gensures that gap links, links directly travelling from left to right encoding simple structural variants, are not penalized.
For further details on these metrics please take a look at the odgi stats command.
We observe on stdout:
#mean_links_length
path in_node_space in_nucleotide_space num_links_considered num_gap_links_not_penalized
all_paths 514.698 4016.92 21870 11116
#sum_of_path_node_distances
path in_node_space in_nucleotide_space nodes nucleotides num_penalties num_penalties_different_orientation
all_paths 1029.84 1076.32 21882 163416 6085 1
Sort the unsorted DRB1-3123 graph in 1D¶
Let’s sort the graph with the 1D PG-SGD algorithm:
odgi sort -i DRB1-3123_unsorted.og --threads 2 -P -Y -o DRB1-3123_sorted.og
-Y selects the PG-SGD algorithm for sorting. This algorithm moves a single pair of nodes at a time, optimizing
the disparity between the layout distance of a node pair and the actual nucleotide distance of a path traversing these
nodes.
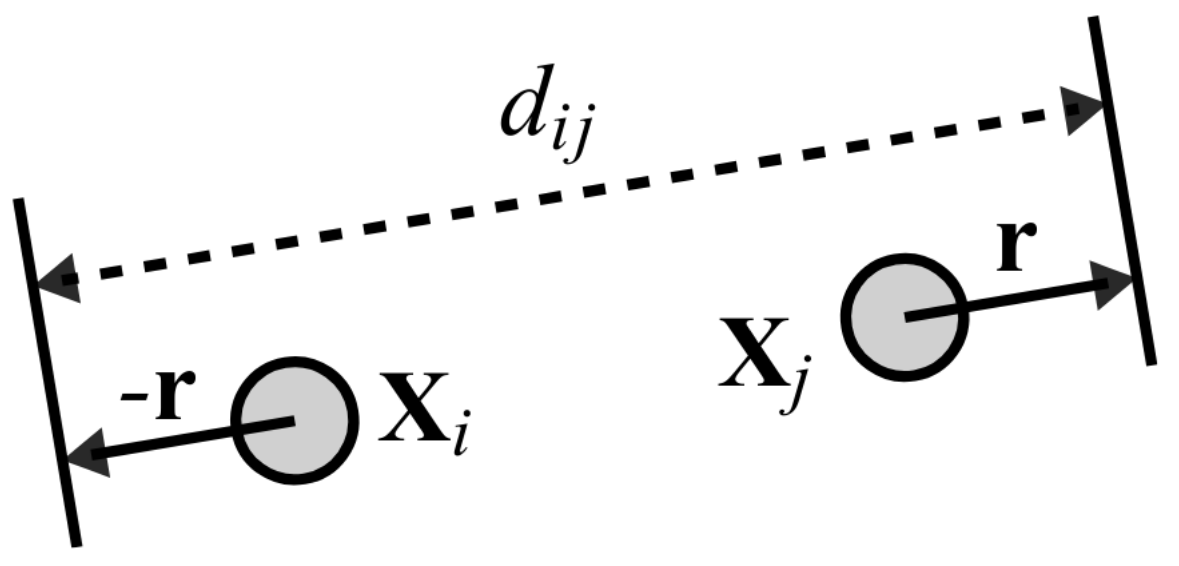
Figure from Zheng et al., IEEE 2019.
The first node Xi of a pair is a uniform path step pick from all nodes.
The second node Xj of a pair is sampled from the same path following a Zipfian distribution.
The path nucleotide distance of the nodes in the pair guides the actual layout distance dij update of these nodes.
The magnitude r of the update depends on the current learning rate of the SGD.
Note
The PG-SGD is not deterministic, because of its Hogwild! approach.
To reproduce the visualization below, the sorted graph can be found under test/DRB1-3123_sorted.og.
Visualize the 1D sorted DRB1-3123 graph¶
odgi viz -i DRB1-3123_sorted.og -o DRB1-3123_sorted.png

The graph lost it’s complexity and is now linear.
1D layout metrics of the sorted DRB1-3123 graph¶
odgi stats -i DRB1-3123_sorted.og -s -d -l -g
This prints to stdout:
#mean_links_length
path in_node_space in_nucleotide_space num_links_considered num_gap_links_not_penalized
all_paths 2.15542 15.0529 21870 9481
#sum_of_path_node_distances
path in_node_space in_nucleotide_space nodes nucleotides num_penalties num_penalties_different_orientation
all_paths 4.66114 4.72171 21882 163416 5948 1
Compare to before, these metrics show that the goodness of the sorting of the graph improved significantly. Great!
2D layout of the unsorted DRB1-3123 graph¶
We want to have a 2D layout of our DRB1-3123 graph:
odgi layout -i DRB1-3123_unsorted.og -o DRB1-3123_unsorted.og.lay -P --threads 2
Drawing the 2D layout of the DRB1-3123 graph¶
Calculate the 2D layout:
odgi draw -i DRB1-3123_unsorted.og -c DRB1-3123_unsorted.og.lay -p DRB1-3123_unsorted.og.lay.png -C -w 50

Interactive visualization with gfaestus¶
odgi layout -i DRB1-3123_unsorted.og -o DRB1-3123_unsorted.og.lay -P --threads 2 -T DRB1-3123_unsorted.og.tsv
gfaestus test/DRB1-3123_unsorted.gfa DRB1-3123_unsorted.og.tsv